/Biological oceanography/Fish
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
status
-
Accredited through the MEDIN partnership, and core-funded by the Department for the Environment, Food and Rural Affairs (Defra) and the Scottish Government, DASSH provides tools and services for the long-term curation, management and publication of marine species and habitats data, within the UK and internationally. Below are a selection of projects, outputs and deliverables that DASSH and the MBA Data Team have been involved in recently. - NE Data Management: DASSH have been contracted by the Marine team at Natural England (NE) to support NE data dissemination. We have been digitising datasets used in Article 17 reporting and helping them input data to Marine Recorder and MEDIN guidelines. In addition, DASSH is running a 2-day workshop with the marine data team in October 2014 on data management and standards. The aims of the workshop are to present MEDIN data guidelines and standards and to run practicals on quality assurance (QA) issues with data, creating MEDIN formatted data, and creation of MEDIN metadata. - MCZ Data Archiving: DASSH staff have been working with Defra, JNCC, Natural England, Cefas and the other MEDIN DAC's in the development and implementation of a strategy for the archiving and dissemination of MCZ survey data. This involves the archives of many terrabytes of data from the survey work undertaken at 127 sites. DASSH is currently working with the other DACs archiving the data from several MCZ sites before taking delivery of the complete survey catalogue. - Non-Natives Data Management: DASSH staff work with other members of the KE team to help deliver the MBA contribution to the GB Non-native Species Information Portal. The data team ensure the validation of records submitted and raise alerts when records of Invasive Non-Native Species of concern and in disseminating information about species distribution via DASSH and the NBN. DASSH staff continue to liaise with organisations to ensure the prompt flow of marine non-native species distribution data to the public domain. The KE team facilitated the identification of two new marine invasive non-native species in 2014 and have subsequently created the identification sheet for these species. Hemigrapsus sanguineus (from volunteer records sent in for identification) and Hemigrapsus takanoi (first recorded by the John Bishop Group survey team). - EMODNet Biology: The Data Team are part of a consortium led by the Flanders Marine Institute (VLIZ) for the biological data component of EMODNet (European Marine Observation and Data Network). The Data Team will lead a work package relating to biological traits and indicator species as identified for Marine Strategy Framework Directive (MSFD) reporting, bringing an additional €130k of funding. - VALMER: The Data Team led a key work package in a £3.7 million (ca. €260k for the MBA) INTERREG project to "Develop, trial and refine methodologies that will be used to quantify and communicate the value (economical, social and environmental) of marine and coastal ecosystem services". The research identified an operational framework to value marine ecosystem services, and which could be used to enhance marine planning and policy decisions.
-

Explore global fisheries and aquaculture. Understand their status and how impacts are being managed. Learn what improvements are underway, and see what actions seafood stakeholders can take to drive sustainability. FishSource is a publicly available online resource about the status of fisheries, fish stocks, and aquaculture. FishSource compiles and summarizes publicly available scientific and technical information and presents it in an easily interpretable form. FishSource was created in 2007 by Sustainable Fisheries Partnership to provide major seafood buyers with up-to-date, impartial, and actionable information on the sustainability of fisheries and the improvements they need to make to become more sustainable. In 2018, information on aquaculture sources was added to the database to provide FishSource users with a more robust tool that covers all types of seafood production. Although the primary intended audience of FishSource is seafood businesses, other audiences - such as academics, researchers, and non-profit organizations - have also become frequent and welcomed users of FishSource. The information on FishSource is primarily developed and updated by a small team of in-house analysts, but their capacity is recognizably insufficient to maintain complete coverage of all global fisheries. As such, profiles may be incomplete or information may be out of date. The seafood industry and external contributors are invited to help fill any gaps that they consider priorities through our Rapid Assessment Program. FishSource always welcome comments on any of our profiles and encourage an open debate on the sources of information used and our interpretation of the data. Our objective is to use only publicly available sources and fully credit those sources, effectively acting as an inventory of information sources on fisheries and aquaculture.
-

OBIS is a global open-access data and information clearing-house on marine biodiversity for science, conservation and sustainable development. VISION: To be the most comprehensive gateway to the world’s ocean biodiversity and biogeographic data and information required to address pressing coastal and world ocean concerns. MISSION: To build and maintain a global alliance that collaborates with scientific communities to facilitate free and open access to, and application of, biodiversity and biogeographic data and information on marine life. More than 20 OBIS nodes around the world connect 500 institutions from 56 countries. Collectively, they have provided over 45 million observations of nearly 120 000 marine species, from Bacteria to Whales, from the surface to 10 900 meters depth, and from the Tropics to the Poles. The datasets are integrated so you can search and map them all seamlessly by species name, higher taxonomic level, geographic area, depth, time and environmental parameters. OBIS emanates from the Census of Marine Life (2000-2010) and was adopted as a project under IOC-UNESCO’s International Oceanographic Data and Information (IODE) programme in 2009. Objectives - Provide world’s largest scientific knowledge base on the diversity, distribution and abundance of all marine organisms in an integrated and standardized format (as a contribution to Aichi biodiversity target 19) - Facilitate the integration of biogeographic information with physical and chemical environmental data, to facilitate climate change studies - Contribute to a concerted global approach to marine biodiversity and ecosystem monitoring, through guidelines on standards and best practices, including globally agreed Essential Ocean Variables, observing plans, and indicators in collaboration with other IOC programs - Support the assessment of the state of marine biological diversity to better inform policymakers, and respond to the needs of regional and global processes such as the UN World Ocean Assessment (WOA) and the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES) - Provide data, information and tools to support the identification of biologically important marine and coastal habitats for the development of marine spatial plans and other area-based management plans (e.g. for the identification of Ecologically or Biologically Significant marine Areas (EBSAs) under the Convention on Biological Diversity. - Increase the institutional and professional capacity in marine biodiversity and ecosystem data collection, management, analysis and reporting tools, as part of IOC’s Ocean Teacher Global Academy (OTGA) - Provide information and guidance on the use of biodiversity data for education and research and provide state of the art services to society including decision-makers - Provide a global platform for international collaboration between national and regional marine biodiversity and ecosystem monitoring programmes, enhancing Member States and global contributions to inter alia, the Global Ocean Observing System (GOOS) and the Global Earth Observing System of Systems (GEOSS)
-
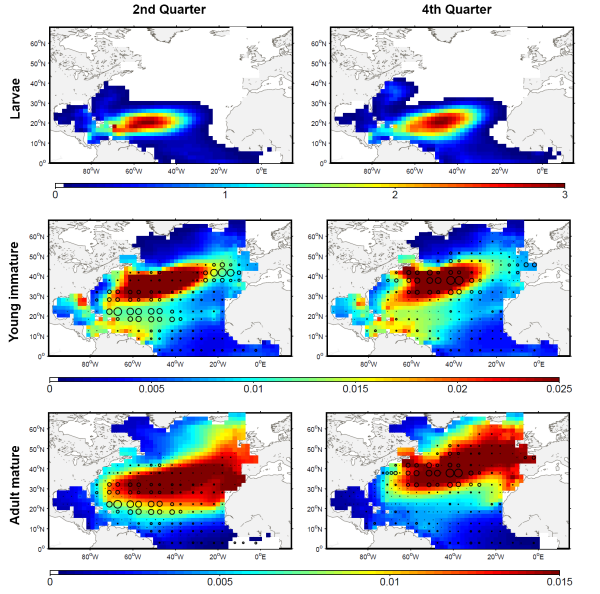
The development of the ecosystem approach and models for the management of ocean marine resources requires easy access to standard validated datasets of historical catch data for the main exploited species. They are used to measure the impact of biomass removal by fisheries and to evaluate the models skills, while the use of standard dataset facilitates models inter-comparison. North Atlantic albacore tuna is exploited all year round by longline and in summer and autumn by surface fisheries and fishery statistics compiled by the International Commission for the Conservation of Atlantic Tunas (ICCAT). Catch and effort with geographical coordinates at monthly spatial resolution of 1° or 5° squares were extracted for this species with a careful definition of fisheries and data screening. In total, thirteen fisheries were defined for the period 1956-2010, with fishing gears longline, troll, mid-water trawl and bait fishing. However, the spatialized catch effort data available in ICCAT database represent a fraction of the entire total catch. Length frequencies of catch were also extracted according to the definition of fisheries above for the period 1956-2010 with a quarterly temporal resolution and spatial resolutions varying from 1°x 1° to 10°x 20°. The resolution used to measure the fish also varies with size-bins of 1, 2 or 5 cm (Fork Length). The screening of data allowed detecting inconsistencies with a relatively large number of samples larger than 150 cm while all studies on the growth of albacore suggest that fish rarely grow up over 130 cm. Therefore, a threshold value of 130 cm has been arbitrarily fixed and all length frequency data above this value removed from the original data set.
-
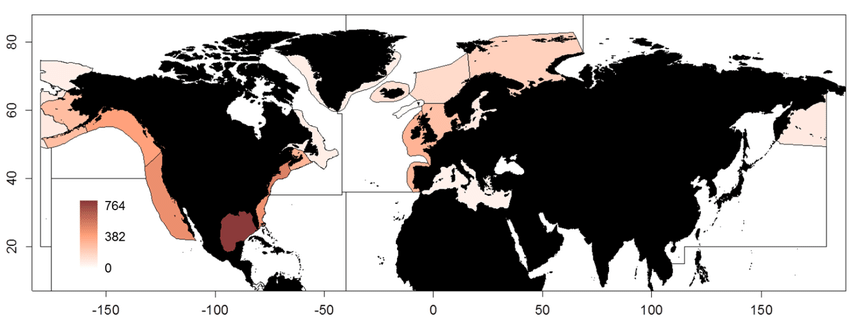
This dataset containing traits of marine fish is based on fish taxa observed during international scientific bottom-trawl surveys regularly conducted in the Northeast Atlantic, Northwest Atlantic and the Northeast Pacific. These scientific surveys target primarily demersal (bottom-dwelling) fish species, but pelagic species are also regularly recorded. The overarching aim of this dataset was to collect information on ecological traits for as many fish taxa as possible and to find area-specific trait values to account for intraspecific variation in traits, especially for widely distributed species. We collected traits for species, genera and families. The majority of trait values were sourced from FishBase (Froese and Pauly, 2019), and have been supplemented with values from the primary literature.
-

The Sea Around Us is a research initiative at The University of British Columbia (located at the Institute for the Oceans and Fisheries, formerly Fisheries Centre) that assesses the impact of fisheries on the marine ecosystems of the world, and offers mitigating solutions to a range of stakeholders. The Sea Around Us was initiated in collaboration with The Pew Charitable Trusts in 1999, and in 2014, the Sea Around Us also began a collaboration with The Paul G. Allen Family Foundation to provide African and Asian countries with more accurate and comprehensive fisheries data. It provides data and analyses through View Data, articles in peer-reviewed journals, and other media (News). We regularly update our products at the scale of countries’ Exclusive Economic Zones, Large Marine Ecosystems, the High Seas and other spatial scales, and as global maps and summaries. It emphasises catch time series starting in 1950, and related series (e.g., landed value and catch by flag state, fishing sector and catch type), and fisheries-related information on every maritime country (e.g., government subsidies, marine biodiversity). Information is also offered on sub-projects, e.g., the historic expansion of fisheries, the performance of Regional Fisheries Management Organizations, or the likely impact of climate change on fisheries. The information and data presented on this website is freely available to any user, granted that its source is acknowledged. We are aware that this information may be incomplete. Please let us know about this via the feedback options available on this website.
-

EMODnet Biology provides three keys services and products to users. 1)The data download toolbox allows users to explore available datasets searching by source, geographical area, and/or time period. Datasets can be narrowed down using a taxonomic criteria, whether by species group (e.g. benthos, fish, algae, pigments) or by both scientific and common name. 2) The data catalogue is the easiest way to access nearly 1000 datasets available through EMODnet Biology. Datasets can be filtered by multiple parameters via the advanced search from taxon, to institute, to geographic region. Each of the resulting datasets then links to a detailed fact sheet containing a link to original data provider, recommended citation, policy and other relevant information. Data Products - EMODnet Biology combines different data from datasets with overlapping geographic scope and produces dynamic maps of selected species abundance. The first products are already available and they focus on species whose data records are most complete and span for a longer term.
-

The abundance of ichthyoplankton in samples from dedicated plankton surveys by Cefas with positional and sample data. Surveys took place off the Western Coast of the UK and Ireland between 1986 and 2004. Series of cruises undertaken to contribute to the estimation of the spawning stock biomass of the western mackerel and horse mackerel stocks by plankton survey. The triennial mackerel egg surveys were begun in 1977 to estimate the SSB of the western mackerel stock. Since 1986 the surveys have also been used to estimate the SSB of horse mackerel. Plankton sampling is undertaken to estimate the egg production and trawling is carried out to estimate the mean fecundity of the mature female fish. Various designs of Gulf VII type samplers have been used with various apertures of nosecones and 270 micron nets. Samplers are now standardised to the 53cm version, fitted with 20cm aperture nosecones. Analysis at Cefas involved separating all fish eggs and larvae from samples. Where possible all eggs were identified. Eggs lacking identifiable features were measured. Where >100 eggs were found, sub-sampling was undertaken. Eggs that were unmeasured were apportioned across the size distribution of measured eggs. All mackerel and horse mackerel eggs were staged.
-
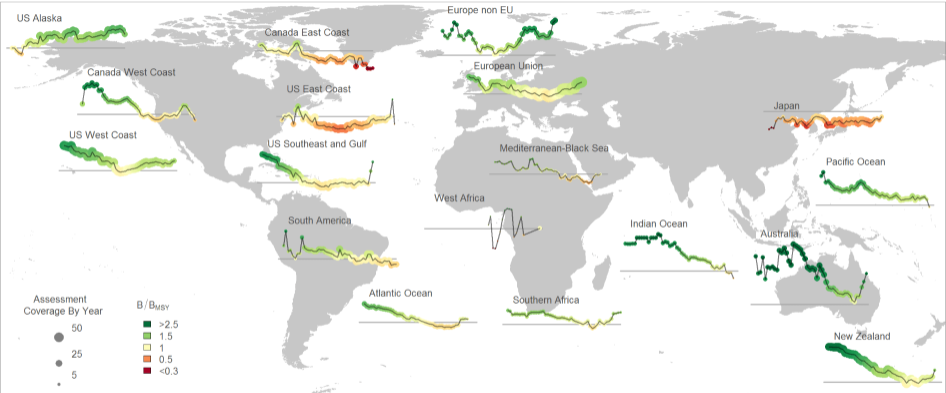
The RAM Legacy Stock Assessment Database is a compilation of stock assessment results for commercially exploited marine populations from around the world. The RAM Legacy Stock Assessment Database is grateful to the many stock assessment scientists whose work this database is based upon and the many collaborators who recorded the assessment model results for inclusion in the RAM Legacy Stock Assessment Database. Since 2011 the RAM Legacy Data base has been hosted and managed at the University of Washington with financial assistance from a consortium of Seattle-based seafood companies and organizations, and from the Walton Family Foundation. Initial development of the database from 2006-2010 was supported by the Census of Marine Life, Canadian Foundation for Innovation, NCEAS, NSERC, the Smith Conservation Research Fellowship, New Jersey Sea Grant, and the National Science Foundation.
 Catalogue PIGMA
Catalogue PIGMA